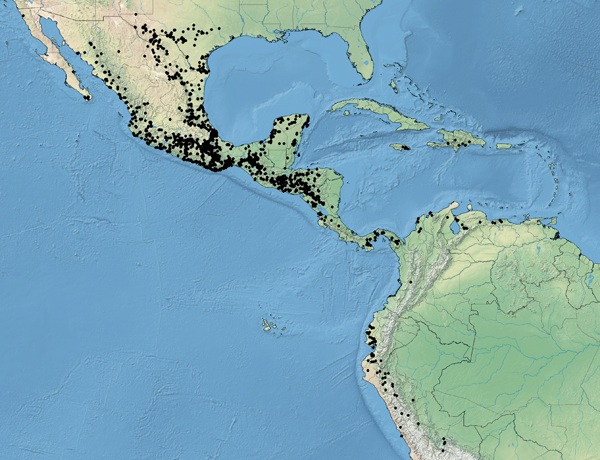
A small genus of 27 species of Neotropical trees
Leucaena (Leguminosae - Mimosoideae) comprises 17 diploid and 5 tetraploid species, six infraspecific taxa, and two named hybrids. They are small to medium-sized trees that grow in seasonally dry tropical forests of Mexico, Central America and northern South America. Species of Leucaena have been the focus of both early indigenous domestication, as a native food plant in Mexico, and modern crop breeding as a fodder tree throughout the tropics.
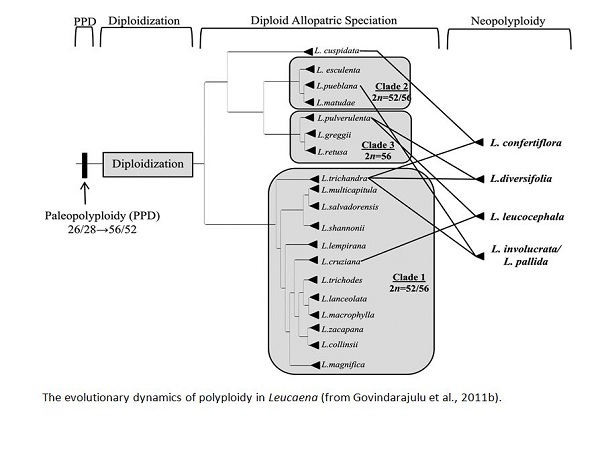
Previous taxonomic work on Leucaena culminated in two major publications - a Taxonomic Monograph of the genus (Hughes, C.E. 1998. Systematic Botany Monographs 55) and a Genetic Resources Handbook (Hughes, C.E. 1998. Oxford Forestry Institute). Recent collaborative research on Leucaena in Oxford and New Mexico has focused on understanding hybrid and polyploid origins using molecular data.
Gene trees, hybrids and polyploid origins
Despite the high frequency of hybridization and polyploidy in plants, significant challenges remain to disentangle reticulation from divergence in plant phylogenies. Recent collaborative research on Leucaena in Oxford and New Mexico has focused on understanding hybrid and polyploid origins using molecular data and a multi-locus gene tree approach. Studies of reticulation demand multiple, densely sampled, highly resolved and robustly supported biparentally inherited gene trees. Much of the work on Leucaena has been devoted to tackling problems posed by lack of informative variation in conventional DNA sequence loci and the occurrence of nrDNA pseudogenes (Hughes et al., 2002; Bailey et al., 2003, 2004) - issues that hinder reconstruction of robust, well-resolved species-level phylogenies for plants. This has involved one of the first studies to thoroughly explore the impacts of nrDNA polymorphism and non-functional pseudogene sequences on phylogeny reconstruction in plants (Hughes et al., 2002; Bailey et al, 2003) and development of a powerful method for identifying pseudogene sequences using phylogenetic trees and bootstrap hypothesis testing (Bailey et al., 2003). We showed that pseudogene sequences are common in nrDNA datasets, which dominate plant species-level phylogenetics, and that their detection is critical for accurate phylogeny reconstruction. To get around the problem of lack of phylogenetic resolution, we developed a novel SCAR-based technique to identify more rapidly-evolving, biparentally-inherited, nuclear DNA sequence loci that can generate gene trees with the required resolution and support (Bailey et al., 2004). Using one of these loci a number of putative hybrids have been investigated (Hughes et al., 2007). Comprehensive sequence data sets for a larger set of SCAR-based nuclear DNA sequence loci are currently under construction by Govindarajulu Rajanikanth, PhD student working with Donovan Bailey. More...